Mini Tools for Gaia data - astroNN.gaia¶
Note
astroNN only contains a limited amount of necessary tools. For a more comprehensive python tool to deal with Gaia data, please refer to Jo Bovy’s gaia_tools
The mission of the GAIA spacecraft is to create a dynamic, three-dimensional map of the Milky Way Galaxy by measuring the distances, positions and proper motion of stars. To do this, the spacecraft employs two telescopes, an imaging system, an instrument for measuring the brightness of stars, and a spectrograph. Launched in 2013, GAIA orbits the Sun at Lagrange point L2, 1.5 million kilometres from Earth. By the end of its five-year mission, GAIA will have mapped well over one billion stars—one percent of the Galactic stellar population.
ESE Gaia satellite: http://sci.esa.int/gaia/
Gaia Data Downloader¶
astroNN Gaia data downloader always act as functions that will return you the path of downloaded file(s),
and download it if it does not exist locally. If the file cannot be found on server, astroNN will generally return
False as the path.
Load Gaia DR2 - Apogee DR14 matches¶
-
astroNN.gaia.gaiadr2_parallax(cuts=True, keepdims=False, offset=False)¶ Load Gaia DR2 - APOGEE DR14 matches, indices corresponds to APOGEE allstar DR14 file
Parameters: - cuts (Union[boolean, float]) – Whether to cut bad data (negative parallax and percentage error more than 20%), or a float to set the threshold
- keepdims (boolean) – Whether to preserve indices the same as APOGEE allstar DR14, no effect when cuts=False, set to -9999 for bad indices when cuts=True keepdims=True
- offset (Union[boolean, float, str]) –
Whether to correction Gaia DR2 zero point offset
- False to assume no offset correction
- True to assume 52.8-4.21(G-12.2)
- ”leungbovydr2” for experimental offset correction
- a float to assume a float offset globally
Returns: numpy array of ra, dec, parallax, parallax_error
Return type: ndarrays
History: 2018-Apr-26 - Written - Henry Leung (University of Toronto)
from astroNN.gaia import gaiadr2_parallax
# To load Gaia DR2 - APOGEE DR14 matches, indices corresponds to APOGEE allstar DR14 file
ra, dec, parallax, parallax_error = gaiadr2_parallax(cuts=True, keepdims=False, offset=False)
Gaia DR1 TGAS Downloader and Loader¶
-
astroNN.gaia.tgas(flag=None)¶ Get path to the Gaia TGAS DR1 files, download if files not found
Returns: List of file path Return type: list History: 2017-Oct-13 - Written - Henry Leung (University of Toronto)
To download TGAS DR1, moreover TGAS is only available in DR1
from astroNN.gaia import tgas
# To download tgas dr1 to GAIA_TOOLS_DATA and it will return the list of path to those files
files_paths = tgas()
To load Gaia TGAS
-
astroNN.gaia.tgas_load(cuts=True)¶ To load useful parameters from multiple TGAS DR1 files
Parameters: cuts (Union[boolean, 0.2]) – Whether to cut bad data (negative parallax and percentage error more than 20%, or a custom cut percentage) Returns: Dictionary of parameters Return type: dict History: 2017-Dec-17 - Written - Henry Leung (University of Toronto)
from astroNN.gaia import tgas_load
# To load the tgas DR1 files and return a dictionary of ra(J2015), dec(J2015), pmra, pmdec, parallax, parallax error, g-band mag
# cuts=True to cut bad data (negative parallax and percentage error more than 20%)
output = tgas_load(cuts=True)
# outout dictionary
output['ra'] # ra(J2015)
output['dec'] # dec(J2015)
output['pmra'] # proper motion in RA
output['pmdec'] # proper motion in DEC
output['parallax'] # parallax
output['parallax_err'] # parallax error
output['gmag'] # g-band mag
Gaia_source DR1 Downloader¶
No plan to support DR2 Gaia Source, please refers to Jo Bovy’s https://github.com/jobovy/gaia_tools
from astroNN.gaia import gaia_source
# To download gaia_source DR1 to GAIA_TOOLS_DATA and it will return the list of path to those files
files_paths = gaia_source(dr=1)
Anderson et al 2017 Improved Parallax from Data-driven Stars Model¶
Anderson2017 is described in here: https://arxiv.org/pdf/1706.05055
Please be advised starting from 26 April 2018, anderson2017 in astroNN is reduced to parallax cross matched with APOGEE DR14 only. If you see this message, anderson2017 in this astroNN version is reduced. Moreover, anderson2017 will be removed in the future
from astroNN.gaia import anderson_2017_parallax
# To load the improved parallax
# Both parallax and para_var is in mas
# cuts=True to cut bad data (negative parallax and percentage error more than 20%)
ra, dec, parallax, para_err = anderson_2017_parallax(cuts=True)
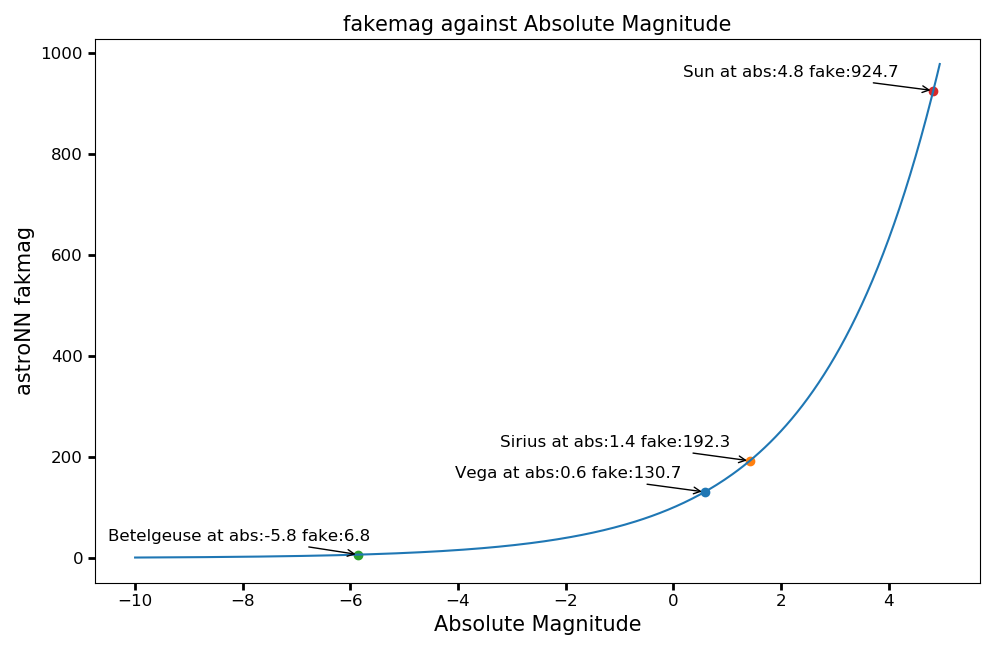
fakemag (dummy scale)¶
fakemag is an astroNN dummy scale primarily used to preserve the gaussian standard error from Gaia. astroNN
always assume there is no error in apparent magnitude measurement.
\(M_\mathrm{fakemag} = \varpi 10^{\frac{1}{5}M_\mathrm{apparent}} = 10^{\frac{1}{5}M_\mathrm{absolute}+2}\), where \(\varpi\) is parallax in mas
You can get a sense of the fakemag scale from the following plot
Coordinates Matching between catalogs using Bovy’s xmatch¶
Coordinates matching between catalogue can be done by xmatch which is just an exact copy from Jo Bovy’s gaia_tools
Here is the documentation of xmatch from Jo Bovy
xmatch(cat1,cat2,maxdist=2, colRA1='RA',colDec1='DEC',epoch1=2000., colRA2='RA',colDec2='DEC',epoch2=2000.,
colpmRA2='pmra',colpmDec2='pmdec', swap=False)
cat1 = First catalog
cat2 = Second catalog
maxdist = (2) maximum distance in arcsec
colRA1 = ('RA') name of the tag in cat1 with the right ascension in degree in cat1 (assumed to be ICRS)
colDec1 = ('DEC') name of the tag in cat1 with the declination in degree in cat1 (assumed to be ICRS)
epoch1 = (2000.) epoch of the coordinates in cat1
colRA2 = ('RA') name of the tag in cat2 with the right ascension in degree in cat2 (assumed to be ICRS)
colDec2 = ('DEC') name of the tag in cat2 with the declination in degree in cat2 (assumed to be ICRS)
epoch2 = (2000.) epoch of the coordinates in cat2
colpmRA2 = ('pmra') name of the tag in cat2 with the proper motion in right ascension in degree in cat2
(assumed to be ICRS; includes cos(Dec)) [only used when epochs are different]
colpmDec2 = ('pmdec') name of the tag in cat2 with the proper motion in declination in degree in cat2
(assumed to be ICRS) [only used when epochs are different]
swap = (False) if False, find closest matches in cat2 for each cat1 source, if False do the opposite (important when one of the catalogs
Here is an example
from astroNN.datasets import xmatch
import numpy as np
# Some coordinates for cat1, J2000.
cat1_ra = np.array([36.,68.,105.,23.,96.,96.])
cat1_dec = np.array([72.,56.,54.,55.,88.,88.])
# Some coordinates for cat2, J2000.
cat2_ra = np.array([23.,56.,222.,96.,245.,68.])
cat2_dec = np.array([36.,68.,82.,88.,26.,56.])
# Using maxdist=2 arcsecond separation threshold, because its default, so not shown here
# Using epoch1=2000. and epoch2=2000., because its default, so not shown here
# because both datasets are J2000., so no need to provide pmra and pmdec which represent proper motion
idx_1, idx_2, sep = xmatch(cat1_ra, cat2_ra, colRA1=cat1_ra, colDec1=cat1_dec, colRA2=cat2_ra, colDec2=cat2_dec, swap=False)
print(idx_1)
>>> [1 4 5]
print(idx_2)
>>> [5 3 3]
print(cat1_ra[idx_1], cat2_ra[idx_2])
>>> [68. 96. 96.], [68. 96. 96.]
# What happens if we swap cat_1 and cat_2
idx_1, idx_2, sep = xmatch(cat2_ra, cat1_ra, colRA1=cat2_ra, colDec1=cat2_dec, colRA2=cat1_ra, colDec2=cat1_dec, swap=False)
print(idx_1)
>>> [3 5]
print(idx_2)
>>> [4 1]
print(cat1_ra[idx_2], cat2_ra[idx_1])
>>> [96. 68.], [96. 68.] # xmatch cant find all the match
# Because we have some repeated index in cat2, we should turn swap=True
idx_1, idx_2, sep = xmatch(cat2_ra, cat1_ra, colRA1=cat2_ra, colDec1=cat2_dec, colRA2=cat1_ra, colDec2=cat1_dec, swap=True)
print(idx_1)
>>> [5 3 3]
print(idx_2)
>>> [1 4 5]
print(cat1_ra[idx_2], cat2_ra[idx_1])
>>> [68. 96. 96.], [68. 96. 96.] # Yea, seems like xmatch found all the matched