APOGEE Spectra with Bayesian Neural Net - ApogeeBCNN
- class astroNN.models.apogee_models.ApogeeBCNN(lr=0.0005, dropout_rate=0.3)[source]
Class for Bayesian convolutional neural network for stellar spectra analysis
- History:
2017-Dec-21 - Written - Henry Leung (University of Toronto)
Although in theory you can feed any 1D data to astroNN neural networks. This tutorial will only focus on spectra analysis.
from astroNN.models import ApogeeBCNN
from astroNN.datasets import H5Loader
# Load the train data from dataset first, x_train is spectra and y_train will be ASPCAP labels
loader = H5Loader('datasets.h5')
loader.load_combined = True
loader.load_err = True
x_train, y_train, x_err, y_err = loader.load()
# And then create an instance of Bayesian Convolutional Neural Network class
bcnn_net = ApogeeBCNN()
# You don't have to specify the task because its 'regression' by default. But if you are doing classification. you can set task='classification'
bcnn_net.task = 'regression'
# Set max_epochs to 10 for a quick result. You should train more epochs normally, especially with dropout
bcnn_net.max_epochs = 10
bcnn_net.train(x_train, y_train, x_err, y_err)
Here is a list of parameter you can set but you can also not set them to use default
ApogeeBCNN.batch_size = 64
ApogeeBCNN.initializer = 'he_normal'
ApogeeBCNN.activation = 'relu'
ApogeeBCNN.num_filters = [2, 4]
ApogeeBCNN.filter_len = 8
ApogeeBCNN.pool_length = 4
ApogeeBCNN.num_hidden = [196, 96]
ApogeeBCNN.max_epochs = 100
ApogeeBCNN.lr = 0.005
ApogeeBCNN.reduce_lr_epsilon = 0.00005
ApogeeBCNN.reduce_lr_min = 0.0000000001
ApogeeBCNN.reduce_lr_patience = 10
ApogeeBCNN.target = 'all'
ApogeeBCNN.l2 = 5e-9
ApogeeBCNN.dropout_rate = 0.2
ApogeeBCNN.length_scale = 0.1 # prior length scale
ApogeeBCNN.input_norm_mode = 3
ApogeeBCNN.labels_norm_mode = 2
Note
You can disable astroNN data normalization via ApogeeBCNN.input_norm_mode=0 as well as ApogeeBCNN.labels_norm_mode=0 and do normalization yourself. But make sure you don’t normalize labels with MAGIC_NUMBER (missing labels).
After the training, you can use bcnn_net in this case and call test method to test the neural network on test data. Or you can load the folder by
from astroNN.models import load_folder
bcnn_net = load_folder('astroNN_0101_run001')
# Load the test data from dataset, x_test is spectra and y_test will be ASPCAP labels
loader2 = H5Loader('datasets.h5')
loader2.load_combined = False
loader2.load_err = False
x_test, y_test = loader2.load()
# pred contains denormalized result aka. ASPCAP labels prediction in this case
# pred_std is a list of uncertainty
# pred_std['total'] is the total uncertainty (standard derivation) which is the sum of all the uncertainty
# pred_std['predictive'] is the predictive uncertainty predicted by bayesian neural net
# pred_std['model'] is the model uncertainty from dropout variational inference
pred, pred_std = bcnn_net.test(x_test)
Since astroNN.models.ApogeeBCNN uses Bayesian deep learning which provides uncertainty analysis features.
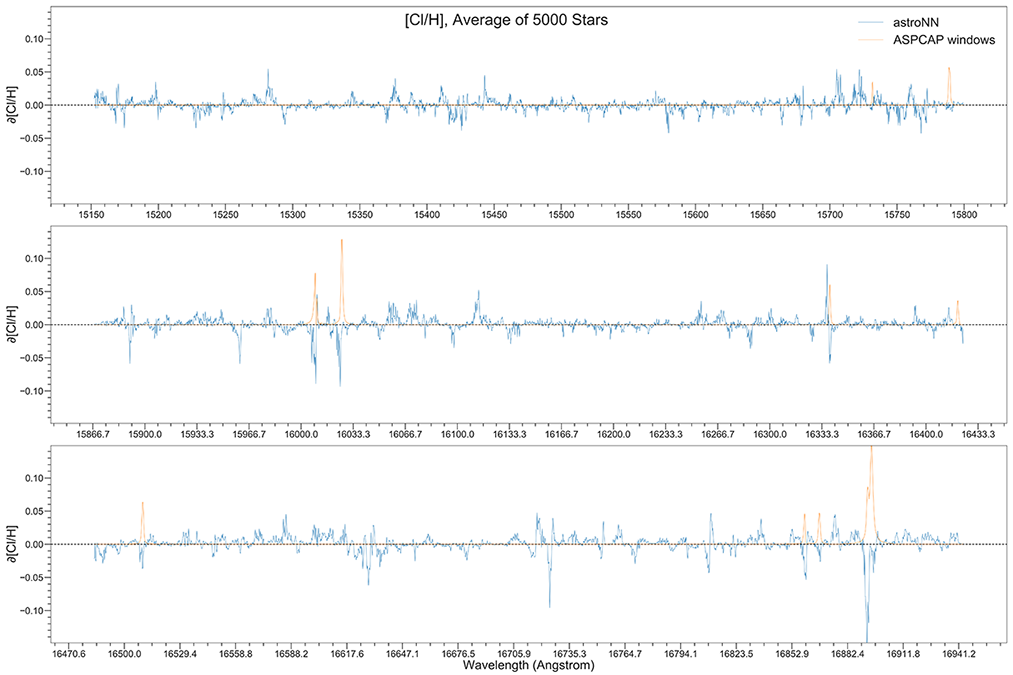
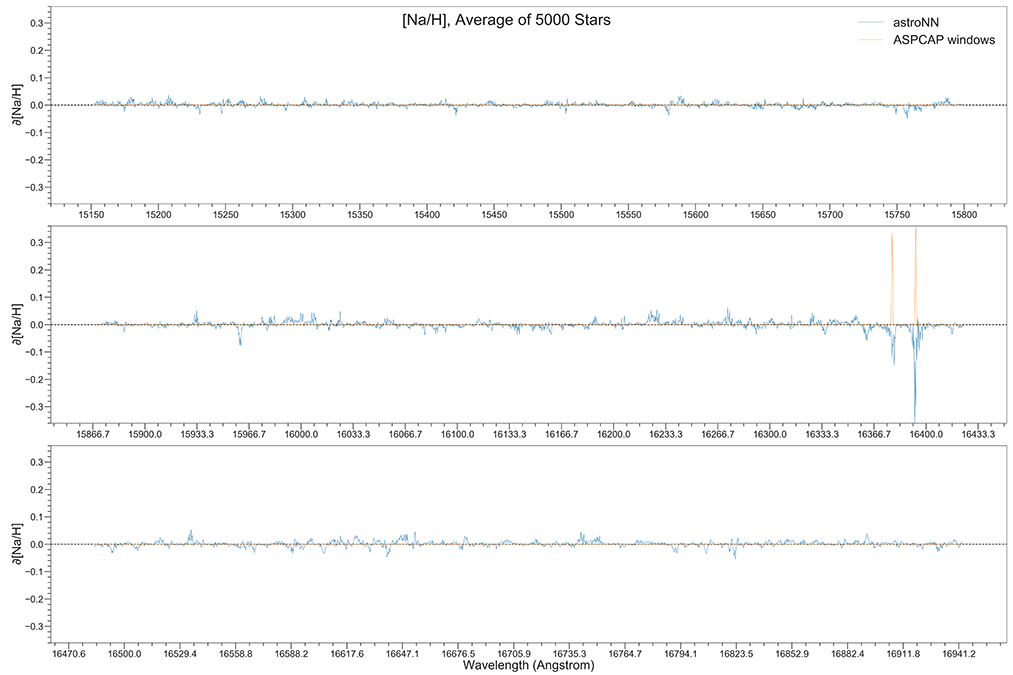
You can calculate jacobian which represents the output derivative to the input and see where those output is sensitive to in inputs.
# Calculate jacobian first
jacobian_array = bcnn_net.jacobian(x_test, mean_output=True)
Note
You can access to Keras model method like model.predict via (in the above tutorial) bcnn_net.keras_model (Example: bcnn_net.keras_model.predict())
ASPCAP Labels Prediction
Internal model identifier for the author: astroNN_0321_run002
Training set (30067 spectra + separate 3340 validation spectra): Starflag=0 and ASPCAPflag=0, 4000<Teff<5500, 200<SNR
Testing set (97723 spectra): Individual Visit of the training spectra, median SNR is around SNR~100
Using astroNN.models.ApogeeBCNN with default hyperparameter
Ground Truth is ASPCAP labels.
Median of residue |
astropy mad_std of residue |
|
|---|---|---|
Al |
-0.003 |
0.042 |
Alpha |
0.000 |
0.013 |
C |
0.003 |
0.032 |
C1 |
0.005 |
0.037 |
Ca |
0.002 |
0.022 |
Co |
-0.005 |
0.071 |
Cr |
-0.001 |
0.031 |
fakemag |
3.314 |
16.727 |
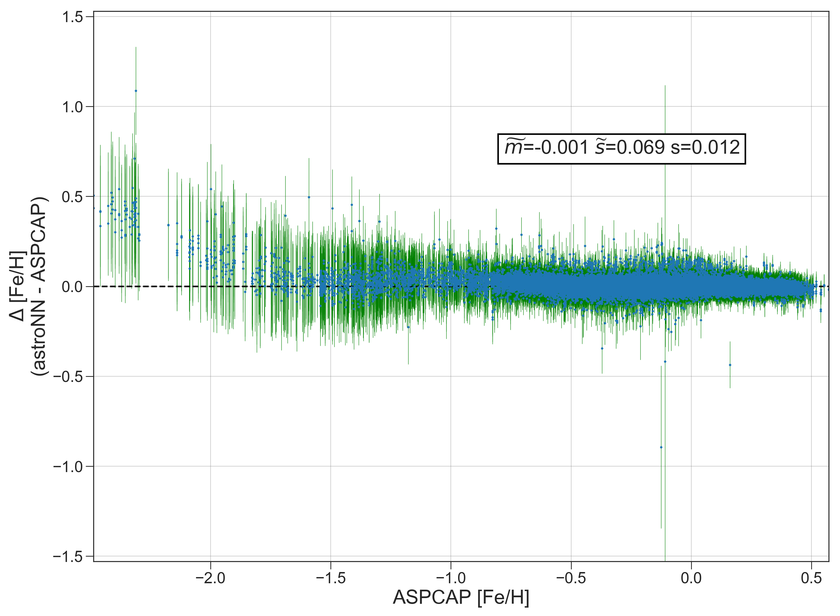
Fe |
0.001 |
0.016 |
K |
-0.001 |
0.032 |
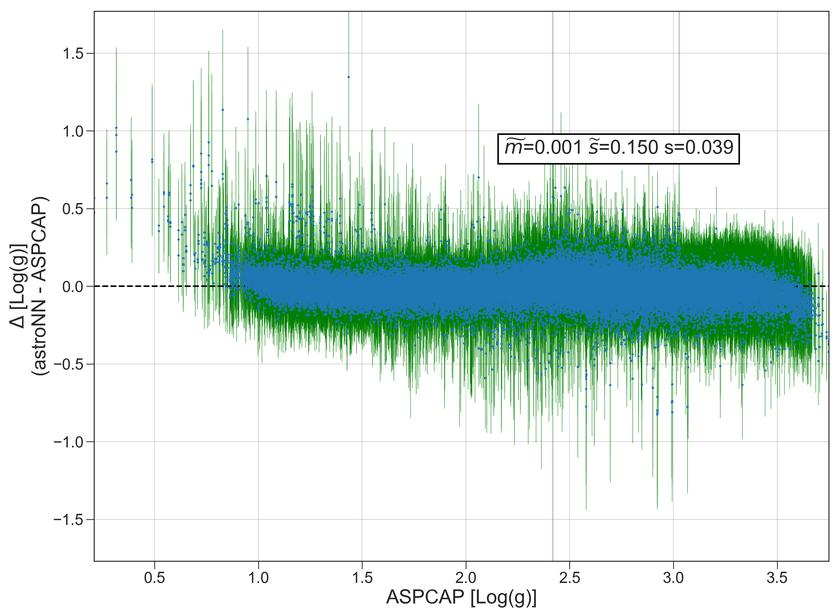
Log(g) |
0.002 |
0.048 |
M |
0.003 |
0.015 |
Mg |
0.001 |
0.021 |
Mn |
0.003 |
0.025 |
N |
-0.002 |
0.037 |
Na |
-0.006 |
0.103 |
Ni |
0.000 |
0.021 |
O |
0.004 |
0.027 |
P |
0.005 |
0.086 |
S |
0.006 |
0.043 |
Si |
0.001 |
0.022 |
Teff |
0.841 |
23.574 |
Ti |
0.002 |
0.032 |
Ti2 |
-0.009 |
0.089 |
V |
-0.002 |
0.059 |
Median Absolute Error of prediction at three different low SNR level.
SNR ~ 20 |
SNR ~ 40 |
SNR ~ 60 |
|
|---|---|---|---|
Al |
0.122 dex |
0.069 dex |
0.046 dex |
Alpha |
0.024 dex |
0.017 dex |
0.014 dex |
C |
0.088 dex |
0.051 dex |
0.037 dex |
C1 |
0.084 dex |
0.054 dex |
0.041 dex |
Ca |
0.069 dex |
0.039 dex |
0.029 dex |
Co |
0.132 dex |
0.104 dex |
0.085 dex |
Cr |
0.082 dex |
0.049 dex |
0.037 dex |
fakemag |
Not Calculated |
Not Calculated |
Not Calculated |
Fe |
0.070 dex |
0.035 dex |
0.024 dex |
K |
0.091 dex |
0.050 dex |
0.037 dex |
Log(g) |
0.152 dex |
0.085 dex |
0.059 dex |
M |
0.067 dex |
0.033 dex |
0.023 dex |
Mg |
0.080 dex |
0.039 dex |
0.026 dex |
Mn |
0.089 dex |
0.050 dex |
0.037 dex |
N |
0.118 dex |
0.067 dex |
0.046 dex |
Na |
0.119 dex |
0.110 dex |
0.099 dex |
Ni |
0.076 dex |
0.039 dex |
0.027 dex |
O |
0.076 dex |
0.046 dex |
0.037 dex |
P |
0.106 dex |
0.082 dex |
0.077 dex |
S |
0.072 dex |
0.052 dex |
0.041 dex |
Si |
0.076 dex |
0.042 dex |
0.024 dex |
Teff |
74.542 K |
41.955 K |
29.271 K |
Ti |
0.080 dex |
0.049 dex |
0.037 dex |
Ti2 |
0.124 dex |
0.099 dex |
0.092 dex |
V |
0.119 dex |
0.080 dex |
0.064 dex |
ASPCAP Labels Prediction with >50% corrupted labels
Internal model identifier for the author: astroNN_0224_run004
Setting is the same as above, but manually corrupt more labels to ensure the modified loss function is working fine
52.5% of the total training labels is corrupted to -9999 (4.6% of the total labels are -9999. from ASPCAP), while testing set is unchanged
Median of residue |
astropy mad_std of residue |
|
|---|---|---|
Al |
0.003 |
0.047 |
Alpha |
0.000 |
0.015 |
C |
0.005 |
0.037 |
C1 |
0.003 |
0.042 |
Ca |
0.002 |
0.025 |
Co |
0.001 |
0.076 |
Cr |
0.000 |
0.033 |
fakemag |
-0.020 |
5.766 |
Fe |
0.001 |
0.020 |
K |
0.001 |
0.035 |
Log(g) |
-0.002 |
0.064 |
M |
0.002 |
0.019 |
Mg |
0.003 |
0.025 |
Mn |
0.003 |
0.030 |
N |
0.001 |
0.043 |
Na |
-0.004 |
0.106 |
Ni |
0.001 |
0.025 |
O |
0.004 |
0.031 |
P |
0.004 |
0.091 |
S |
0.006 |
0.045 |
Si |
0.001 |
0.026 |
Teff |
-0.405 |
31.222 |
Ti |
0.003 |
0.035 |
Ti2 |
-0.012 |
0.092 |
V |
0.002 |
0.063 |
ASPCAP Labels Prediction with limited amount of data
Internal model identifier for the author: astroNN_0401_run001
Setting is the same including the neural network, but the number of training data is limited to 5000 (4500 of them is for training, 500 validation), validation set is completely separated. Testing set is the same without any limitation.
Median of residue |
astropy mad_std of residue |
|
|---|---|---|
Al |
-0.002 |
0.051 |
Alpha |
0.001 |
0.017 |
C |
-0.002 |
0.040 |
C1 |
-0.003 |
0.046 |
Ca |
-0.003 |
0.027 |
Co |
-0.006 |
0.080 |
Cr |
0.000 |
0.036 |
fakemag |
18.798 |
30.687 |
Fe |
-0.004 |
0.022 |
K |
-0.003 |
0.038 |
Log(g) |
-0.005 |
0.064 |
M |
-0.004 |
0.020 |
Mg |
-0.002 |
0.026 |
Mn |
-0.002 |
0.033 |
N |
-0.003 |
0.053 |
Na |
-0.026 |
0.121 |
Ni |
-0.003 |
0.026 |
O |
-0.003 |
0.033 |
P |
0.001 |
0.097 |
S |
-0.003 |
0.047 |
Si |
-0.003 |
0.028 |
Teff |
-1.348 |
33.202 |
Ti |
-0.004 |
0.037 |
Ti2 |
-0.017 |
0.097 |
V |
-0.005 |
0.065 |
Example Plots using aspcap_residue_plot
Example Plots using jacobian